Deep sequence models tend to memorize geometrically; it is unclear why.

Deep sequence models tend to memorize geometrically; it is unclear why.
[Preprint: ArXiv]
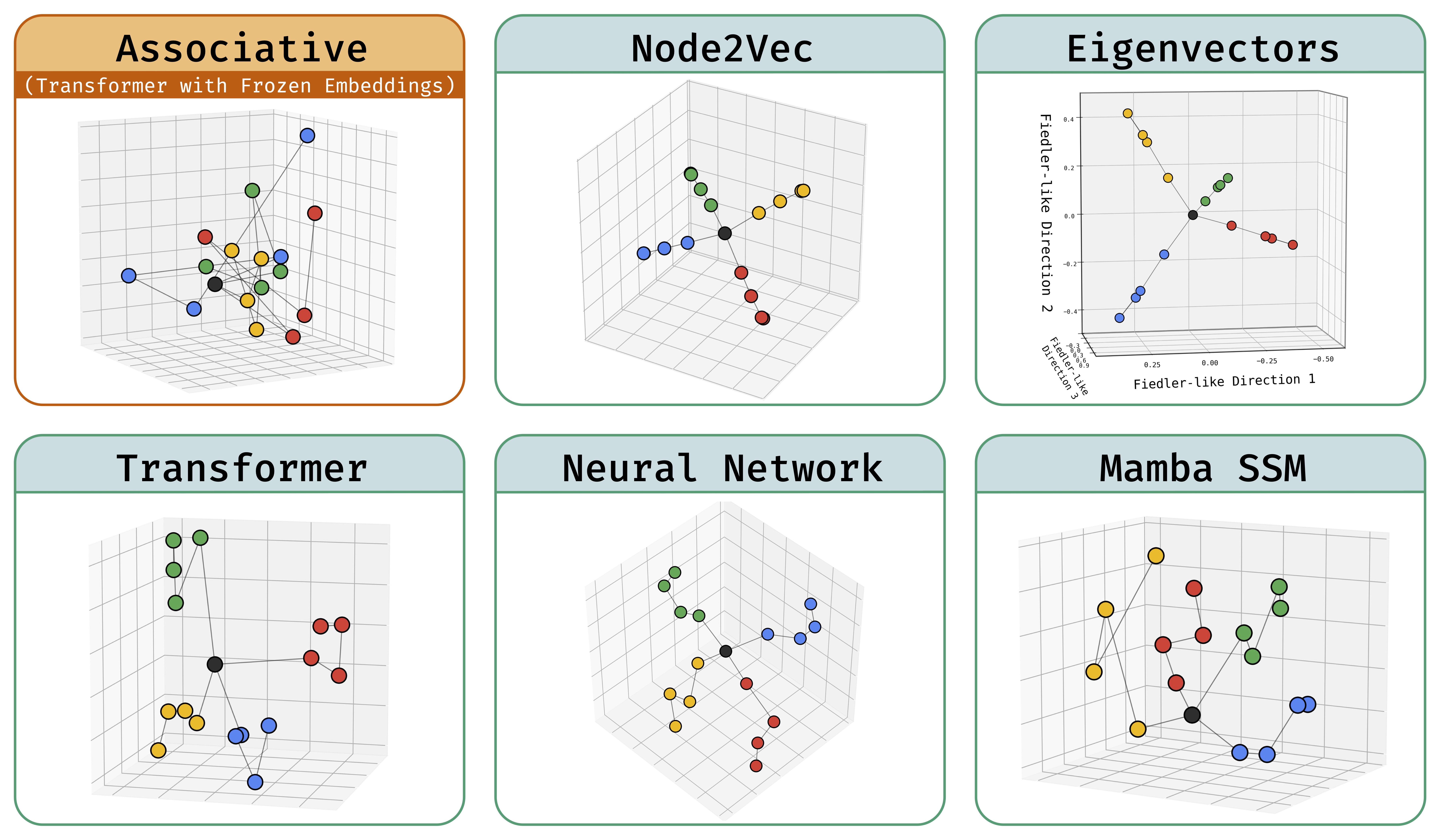
Abstract: Deep sequence models are said to store atomic facts predominantly in the form of associative memory: a brute-force lookup of co-occurring entities. We identify a dramatically different form of storage of atomic facts that we term as geometric memory. Here, the model has synthesized embeddings encoding novel global relationships between all entities, including ones that do not co-occur in training. Such storage is powerful: for instance, we show how it transforms a hard reasoning task involving an ℓ-fold composition into an easy-to-learn 1-step navigation task.
From this phenomenon, we extract fundamental aspects of neural embedding geometries that are hard to explain. We argue that the rise of such a geometry, as against a lookup of local associations, cannot be straightforwardly attributed to typical supervisory, architectural, or optimizational pressures. Counterintuitively, a geometry is learned even when it is more complex than the brute-force lookup.
Then, by analyzing a connection to Node2Vec, we demonstrate how the geometry stems from a spectral bias that -- in contrast to prevailing theories -- indeed arises naturally despite the lack of various pressures. This analysis also points out to practitioners a visible headroom to make Transformer memory more strongly geometric. We hope the geometric view of parametric memory encourages revisiting the default intuitions that guide researchers in areas like knowledge acquisition, capacity, discovery, and unlearning.